Setting up rgbif
rgbif enables programmatic access to GBIF mediated data via its API.
Installing rgbif
Before we can use the R package, we need to install it. In the simplest way this can be done with the install.packages(...) command:
install.packages("rgbif")
Following successful installation of rgbif, we simply need to load it into our R session to make its functionality available to use via the library(...) command:
library(rgbif)
rgbif functionality.
I strongly suggest never installing and loading packages this way in a script that is meant to be reproducible. Using the install.packages(...) command forces installation of an R package irrespective of whether it is already installed or not which leads to three distinct issues:
- Already installed versions of
Rpackages are overwritten potentially breaking pre-existing scripts. - Package installation time is wasted time if packages are already installed ion the first place.
- Package installation is dependent on internet connection making offline sourcing of scripts impossible.
For a user-defined function that avoids these issues, click here:
Below, I create a user defined function called `install.load.package(...)`. This function takes as an input the name of an `R` package just like the `install.packages(...)` function does. Instead of simply forcing package installation, however, `install.load.package(...)` checks whether the package in question is already installed and only carries out installation if it isn't. Before concluding, `install.load.package(...)` loads the package.## Custom install & load function
install.load.package <- function(x) {
if (!require(x, character.only = TRUE)) {
install.packages(x, repos = "http://cran.us.r-project.org")
}
require(x, character.only = TRUE)
}
To further streamline package installation and loading, we can use sapply(...) to execute the install.load.package(...) function sequentially for all the packages we desire or require:
## names of packages we want installed (if not installed yet) and loaded
package_vec <- c(
"rgbif"
)
## executing install & load for each package
sapply(package_vec, install.load.package)
## rgbif
## TRUE
GBIF Account
rgbif functionality can be used without registering an account at
GBIF. However, doing so imposes some crucial limitations:
- Downloads are capped at 100,000 records per download
- Once downloaded, GBIF mediated data needs to be queried again for re-download
- Accrediting and referencing GBIF mediated data becomes difficult
Having a GBIF account and using it for download querying resolves all these limitations. So, let’s get you registered with GBIF.
Opening an Account at GBIF
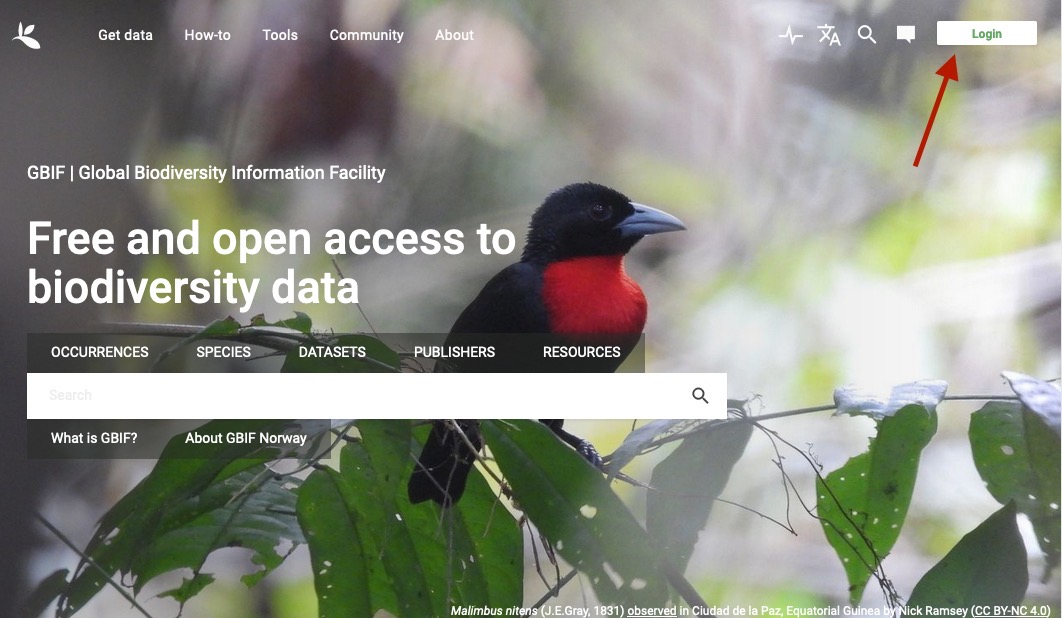
First, navigate to
gbif.org and click the Login button:
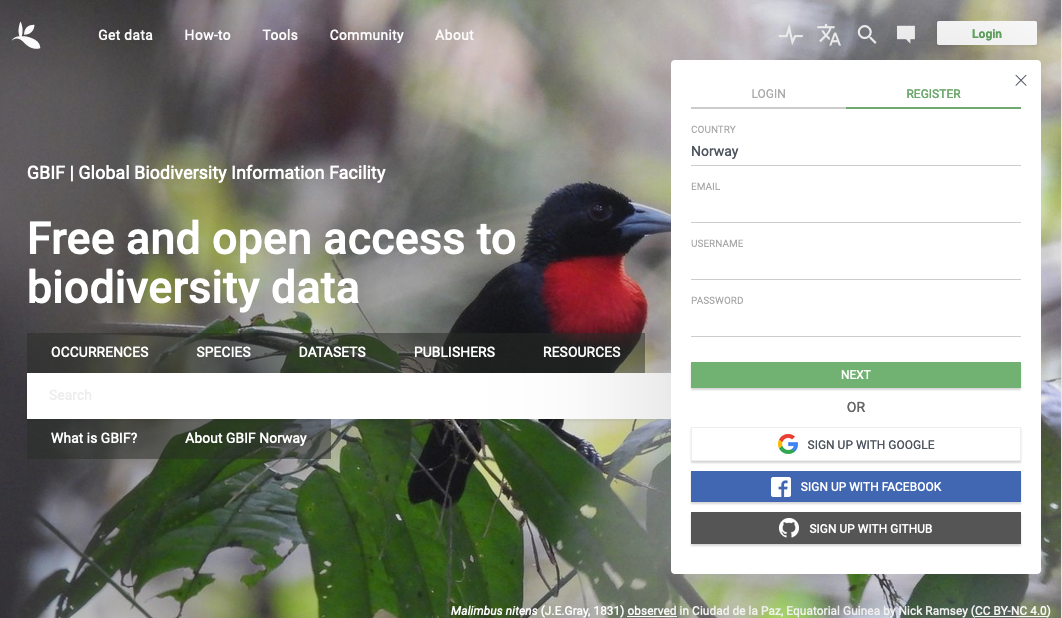
Chose your preferred method of registering your account:
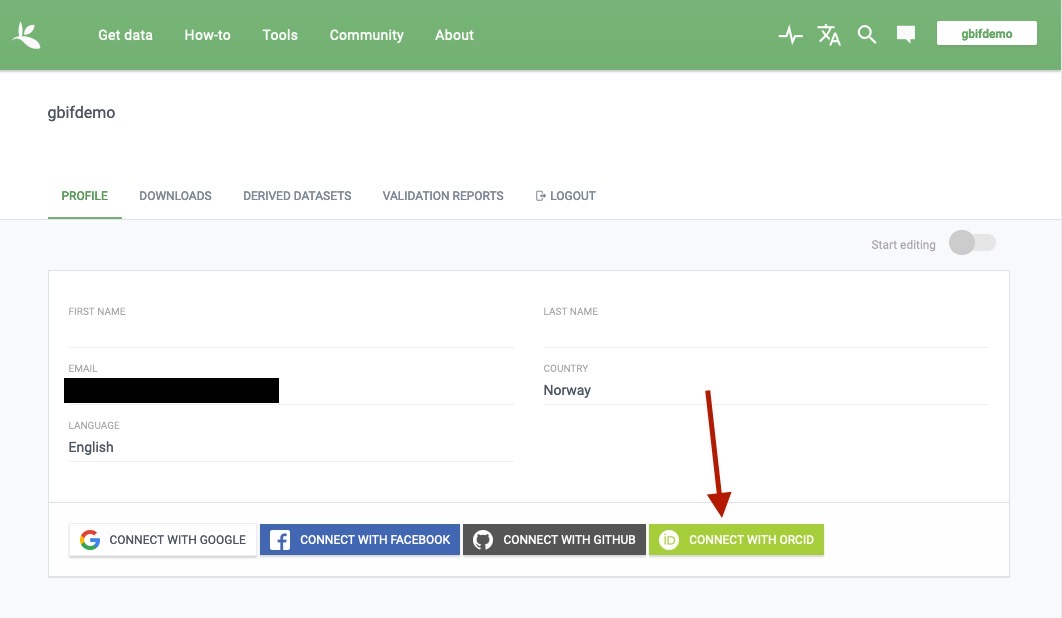
Connect your
ORCID. This step is not strictly necessary, but will make subsequent logins very straightforward and streamline citation and data use tracking:
Registering your GBIF Account in R
Lastly, we need to tell your R session about your GBIF credentials. This can either be done for each individual function call executed from rgbif to the GBIF API, or set once per R session. I prefer the latter, so let’s register your GBIF credentials as follows:
options(gbif_user = "my gbif username")
options(gbif_email = "my registred gbif e-mail")
options(gbif_pwd = "my gbif password")
rgbif.