Discovering Data with rgbif
Preamble, Package-Loading, and GBIF API Credential Registering (click here):
## Custom install & load function
install.load.package <- function(x) {
if (!require(x, character.only = TRUE)) {
install.packages(x, repos = "http://cran.us.r-project.org")
}
require(x, character.only = TRUE)
}
## names of packages we want installed (if not installed yet) and loaded
package_vec <- c(
"rgbif",
"knitr", # for rmarkdown table visualisations
"rnaturalearth", # for shapefile access
"sf", # for transforming polygons into wkt format
"ggplot2", # for visualisations
"networkD3", # for sankey plots
"htmlwidgets", # for sankey inclusion in website
"leaflet", # for map visualisation
"pbapply" # for apply functions with progress bars which I use in hidden code chunks
)
## executing install & load for each package
sapply(package_vec, install.load.package)
## rgbif knitr rnaturalearth sf ggplot2 networkD3
## TRUE TRUE TRUE TRUE TRUE TRUE
## htmlwidgets leaflet pbapply
## TRUE TRUE TRUE
options(gbif_user = "my gbif username")
options(gbif_email = "my registred gbif e-mail")
options(gbif_pwd = "my gbif password")
First, we need to register the correct usageKey for Lagopus muta:
sp_name <- "Lagopus muta"
sp_backbone <- name_backbone(name = sp_name)
sp_key <- sp_backbone$usageKey
With usageKey in hand, we are now ready to discover relevant data for our study needs. Doing so is a complex task mediated through rigurous metadata standards on GBIF.
Data on GBIF is indexed according to the Darwin Core Standard. Any discovery of data can be augmented by matching terms of the Darwin Core with desired subset characteristics. Here, I will show how we can build an increasingly complex query for Lagopus muta records.
At base, we will start with the functions occc_count(...) and occ_search(...):
occ_count(taxonKey = sp_key)
## [1] 107599
occ_search(taxonKey = sp_key, limit = 0)$meta$count
## [1] 107599
Using both of these, we obtain the same output of
occ_search(...) function as strings with semicolons separating desired characteristics (e.g., year = "2000;2001").
Spatial Limitation
By Country Code
First, let’s limit our search by a specific region. In this case, we are interested in occurrences of Lagopus muta across Norway. Countries are indexed according to ISO2 names (two-letter codes, see enumeration_country()):
occ_NO <- occ_search(taxonKey = sp_key, country = "NO")
occ_NO$meta$count
## [1] 17028
Here is how Lagopus muta records are distributed across countries according to GBIF on 2024-11-15:
Click here for code necessary to create the figure below.
occ_countries <- occ_count(
taxonKey = sp_key,
facet = c("country"),
facetLimit = nrow(enumeration_country())
)
occ_countries <- cbind(
occ_countries,
data.frame(
GBIF = "GBIF",
Countries = enumeration_country()$title[
match(occ_countries$country, enumeration_country()$iso2)
]
)
)
occ_countries$Records <- as.numeric(occ_countries$count)
occ_countries <- occ_countries[occ_countries$Records != 0, ]
occ_countries <- occ_countries[, -1]
plot_df <- rbind(
occ_countries[order(occ_countries$Records, decreasing = TRUE)[1:10], -1],
data.frame(
GBIF = "GBIF",
Countries = "Others",
Records = sum(occ_countries$Records[order(occ_countries$Records, decreasing = TRUE)[-1:-10]])
)
)
plot_df$Countries[plot_df$Countries == "United Kingdom of Great Britain and Northern Ireland"] <- "United Kingdom"
alluvial_df <- plot_df
my_color <- 'd3.scaleOrdinal() .range(["#FDE725FF","#1F9E89FF"])'
links <- alluvial_df
colnames(links) <- c("source", "target", "value")
nodes <- data.frame(
name = c("GBIF", alluvial_df$Countries),
group = c("a", rep("b", nrow(alluvial_df)))
)
links$IDsource <- match(links$source, nodes$name) - 1
links$IDtarget <- match(links$target, nodes$name) - 1
SN <- sankeyNetwork(
Links = links, Nodes = nodes,
Source = "IDsource", Target = "IDtarget",
Value = "value", NodeID = "name",
colourScale = my_color, NodeGroup = "group",
sinksRight = FALSE, nodeWidth = 40, fontSize = 13, nodePadding = 5
)
networkD3::saveNetwork(network = SN, file = "SankeyCountry.html", selfcontained = TRUE)
As it turns out, roughly 15.83% of Lagopus muta records mediated by GBIF fall within Norway.
By Shapefile / Polygon
Oftentimes, you won’t be interested in occurrences according to a specific country, but rather a specific area on Earth as identified through a shapefile. To demonstrate data discovery by shapefile, let’s obtain the shapefile for Norway from naturalearth:
NO_shp <- rnaturalearth::ne_countries(country = "Norway", scale = "small", returnclass = "sf")[, 1] # I am extracting only the first column for ease of use, I don't need additional information
This shapefile subsequently looks like this:
Click here for code necessary to create the figure below.
shape_leaflet <- leaflet(NO_shp) %>%
addTiles() %>%
addPolygons(col = "red")
saveWidget(shape_leaflet, "leaflet_shape.html", selfcontained = TRUE)
Let’s focus on just continental Norway:
NO_shp <- st_crop(NO_shp, xmin = 4.98, xmax = 31.3, ymin = 58, ymax = 71.14)
Click here for code necessary to create the figure below.
shape_leaflet <- leaflet(NO_shp) %>%
addTiles() %>%
addPolygons(col = "red")
saveWidget(shape_leaflet, "leaflet_shapeCrop.html", selfcontained = TRUE)
Unfortunately, rgbif prefers to be told about shapefiles in Well Known Text (WKT) format so we need to reformat our polygon data frame obtained above. We do so like such:
NO_wkt <- st_as_text(st_geometry(NO_shp))
We can now pass this information into the occ_search(...) function using the geometry argument:
occ_wkt <- occ_search(taxonKey = sp_key, geometry = NO_wkt)
occ_wkt$meta$count
## [1] 14752
Finally, we find that there are fewer records available when using the shapefile for data discovery. Why would that be? In this case, you will find that we are using a pretty coarse shapefile for Norway which probably cuts off some obersvations of Lagopus muta that do belong rightfully into Norway.
When searching for data by country code (or continent code for that matter), the returned data need not necessarily contain coordinates so long as a record is assigned to the relevant country code. While all records whose coordinates fall within a certain country are assigned the corresponding country code, not all records with a country code have coordinates attached. Additionally, the polygon defintion used by GBIF may be different to the one used by naturalearth.
By Time-Frame
Single-Year
When interested in a single year or month of records being obtained, we can use the year and month arguments, respectively. Both take numeric inputs. Here, we are just looking at occurrences of Lagopus muta throughout the year 2020:
occ_year <- occ_search(
taxonKey = sp_key,
country = "NO",
year = 2020
)
occ_year$meta$count
## [1] 1031
Time-Window
When investigating long-term trends and patterns of biodiversity, we are rarely concerned with a single year or month, but rather time-windows of data. These are also discovered using the year and month arguments. In addition to specifying strings with semicolons separating years, we can alternatively also specify a sequence of integers:
occ_window <- occ_search(
taxonKey = sp_key,
country = "NO",
year = 2000:2020
)
This returns a list of discovered data with each list element corresponding to one year worth of data. To sum up how many records are available in the time-window we thus need to run the following:
sum(unlist(lapply(occ_window, FUN = function(x) {
x$meta$count
})))
## [1] 10866
Using the occ_count(...) is easier in this example:
occ_count(
taxonKey = sp_key,
country = "NO",
year = "2000,2020"
)
## [1] 10866
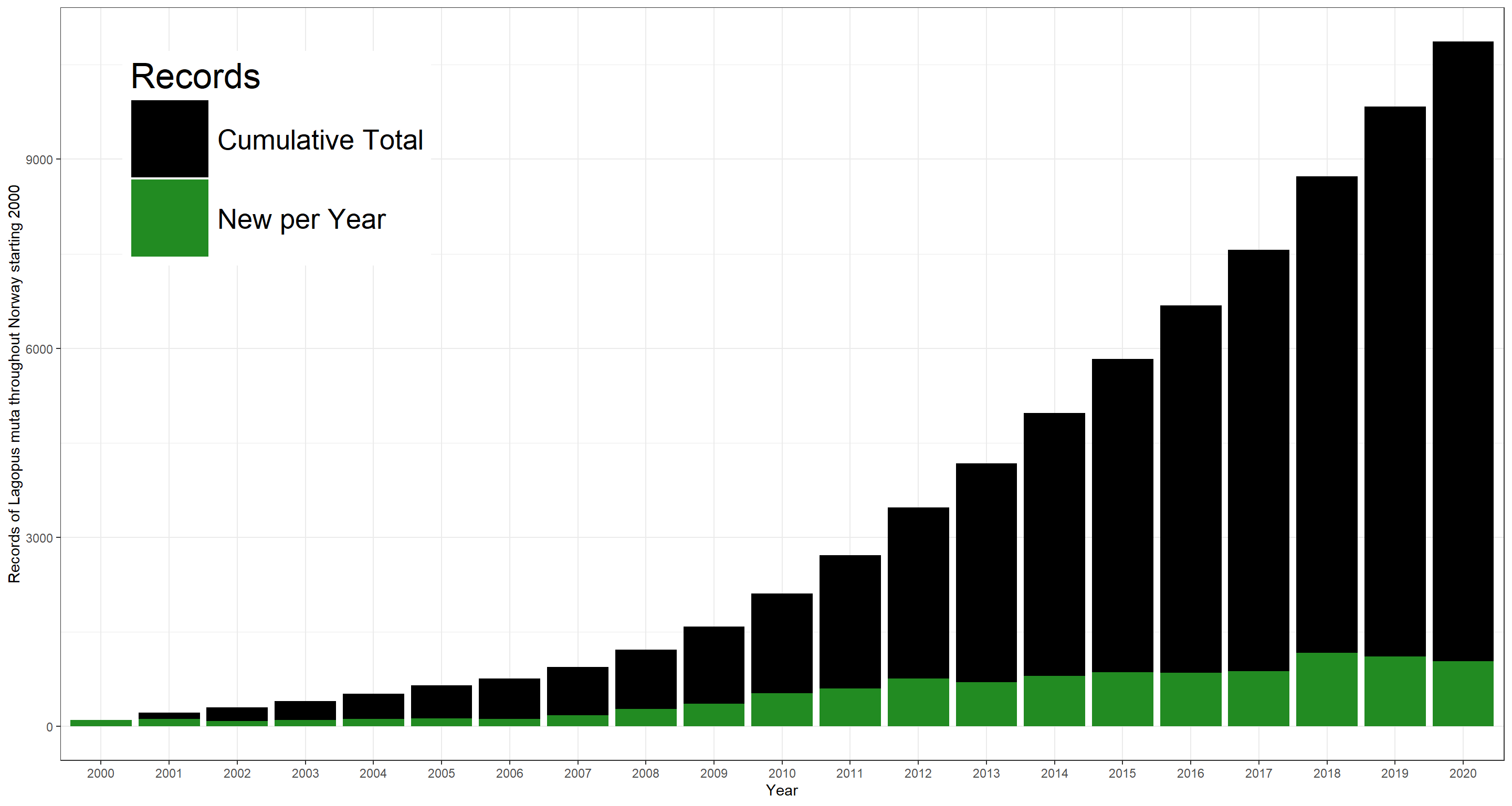
Through time, the number of records develops like this:
Click here for code necessary to create the figure below.
plot_df <- data.frame(
Year = 2000:2020,
Records = unlist(lapply(occ_window, FUN = function(x) {
x$meta$count
})),
Cumulative = cumsum(unlist(lapply(occ_window, FUN = function(x) {
x$meta$count
})))
)
ggplot(data = plot_df, aes(x = as.factor(Year), y = Cumulative)) +
geom_bar(stat = "identity", aes(fill = "black")) +
geom_bar(stat = "identity", aes(y = Records, fill = "forestgreen")) +
theme_bw() +
scale_fill_manual(
name = "Records",
values = c("black" = "black", "forestgreen" = "forestgreen"), labels = c("Cumulative Total", "New per Year")
) +
theme(legend.position = c(0.15, 0.8), legend.key.size = unit(2, "cm"), legend.text = element_text(size = 20), legend.title = element_text(size = 25)) +
labs(x = "Year", y = "Records of Lagopus muta throughout Norway starting 2000")
By Basis of Record
Human Observation
First, let’s look at Lagopus muta occurrences identified by human observation:
occ_human <- occ_search(
taxonKey = sp_key,
country = "NO",
year = 2020,
basisOfRecord = "HUMAN_OBSERVATION"
)
occ_human$meta$count
## [1] 1021
These account for 99.03% of all Lagopus muta observations in Norway throughout the year 2020. So, what might be the basis of record for the remaining 10 records?
Preserved Specimen
As it turns out, the remaining 10 records are based on preserved specimen:
occ_preserved <- occ_search(
taxonKey = sp_key,
country = "NO",
year = 2020,
basisOfRecord = "PRESERVED_SPECIMEN"
)
occ_preserved$meta$count
## [1] 2
These are probably not very useful for many ecological applications which would rather focus on observations of specimen directly.
Other Basis' of Record
There are additional characteristics for basis of record indexing. These are:
- “FOSSIL_SPECIMEN”.
- “HUMAN_OBSERVATION”.
- “MATERIAL_CITATION”.
- “MATERIAL_SAMPLE”.
- “LIVING_SPECIMEN”.
- “MACHINE_OBSERVATION”.
- “OBSERVATION”.
- “PRESERVED_SPECIMEN”.
- “OCCURRENCE”.
Per country containing records of Lagopus muta, the data split ends up looking like this:
Click here for code necessary to create the figure below.
## query basis of record per country
basis_occ <- data.frame(basisofRecord = c(
"FOSSIL_SPECIMEN", "HUMAN_OBSERVATION", "MATERIAL_CITATION", "MATERIAL_SAMPLE",
"LIVING_SPECIMEN", "MACHINE_OBSERVATION", "OBSERVATION", "PRESERVED_SPECIMEN", "OCCURRENCE"
))
basis_occ <- expand.grid(basis_occ$basisofRecord, enumeration_country()$"iso2")
basis_occ$Records <- pbapply(basis_occ, MARGIN = 1, FUN = function(x) {
occ_search(
taxonKey = sp_key,
country = x[2],
basisOfRecord = x[1]
)$meta$count
})
basis_occ$Country <- enumeration_country()$"title"[match(basis_occ$Var2, enumeration_country()$"iso2")]
basis_occ$Country[basis_occ$Country == "United Kingdom of Great Britain and Northern Ireland"] <- "United Kingdom"
## build summary data frame
links2 <- expand.grid(links$target, unique(basis_occ$Var1))
colnames(links2) <- c("source", "target")
links2$value <- 0
links2$value <- unlist(apply(links2, MARGIN = 1, FUN = function(x) {
# print(x)
if (x[1] == "Others") {
aggregate(
data = basis_occ[!(basis_occ$Country %in% nodes$name) & basis_occ$Var1 %in% as.character(x[2]), ],
x = Records ~ 1, FUN = sum
)
} else {
aggr_df <- basis_occ[basis_occ$Country %in% x[1] & basis_occ$Var1 %in% as.character(x[2]), ]
if (nrow(aggr_df) == 0) {
0
} else {
aggregate(
data = aggr_df,
x = Records ~ 1, FUN = sum
)
}
}
}))
## build links and nodes
links_plot <- rbind(links[, 1:3], links2)
links_plot <- links_plot[links_plot$value != 0, ]
nodes_plot <- rbind(
nodes,
data.frame(
name = unique(basis_occ$Var1),
group = "c"
)
)
nodes_plot <- nodes_plot[nodes_plot$name %in% c(links_plot$source, links_plot$target), ]
links_plot$IDsource <- match(links_plot$source, nodes_plot$name) - 1
links_plot$IDtarget <- match(links_plot$target, nodes_plot$name) - 1
## make sankey
my_color <- 'd3.scaleOrdinal() .range(["#FDE725FF","#1F9E89FF","#440154FF"])'
SN <- sankeyNetwork(
Links = links_plot, Nodes = nodes_plot,
Source = "IDsource", Target = "IDtarget",
Value = "value", NodeID = "name",
colourScale = my_color, NodeGroup = "group",
sinksRight = FALSE, nodeWidth = 40, fontSize = 13, nodePadding = 5, margin = 2
)
networkD3::saveNetwork(network = SN, file = "SankeyBasis.html", selfcontained = TRUE)
As should be plain to see from the list above, there exists some ambiguity in regards to which basis of record may apply to any single occurrence record. It is thus always best to carefully examine on which basis of record research projects should be based.
Occurrence Status
To avoid erroneous use of GBIF-mediated data, it is thus always necessary to be precise about what is understood by “occurrence”. This can be controlled with the ocurrenceStatus argument.
Present
First, we ought to look at which occurrences actually report presence of our target organism:
occ_present <- occ_search(
taxonKey = sp_key,
country = "NO",
year = 2020,
basisOfRecord = "HUMAN_OBSERVATION",
occurrenceStatus = "PRESENT"
)
occ_present$meta$count
## [1] 1021
Well, that is all of them.
Absent
Therefore, we should find 0 records reporting absences given our additional limiting characteristics for data discovery:
occ_absent <- occ_search(
taxonKey = sp_key,
country = "NO",
year = 2020,
basisOfRecord = "HUMAN_OBSERVATION",
occurrenceStatus = "ABSENT"
)
occ_absent$meta$count
## [1] 0
That is indeed true.
Are there even any records of absences of Lagopus muta? Let’s check:
occ_absent <- occ_search(
taxonKey = sp_key,
occurrenceStatus = "ABSENT"
)
occ_absent$meta$count
## [1] 2421
Yes, there are, but there are far fewer reported absences than presences.
Let me visualise this one final time on a country-by-country basis:
Click here for code necessary to create the figure below.
## query basis of record per country
basis_occ <- expand.grid(
c("PRESENT", "ABSENT"),
c(
"FOSSIL_SPECIMEN", "HUMAN_OBSERVATION", "MATERIAL_CITATION", "MATERIAL_SAMPLE",
"LIVING_SPECIMEN", "MACHINE_OBSERVATION", "OBSERVATION", "PRESERVED_SPECIMEN", "OCCURRENCE"
)
)
basis_occ$Records <- pbapply(basis_occ, MARGIN = 1, FUN = function(x) {
occ_search(
taxonKey = sp_key,
basisOfRecord = x[2],
occurrenceStatus = x[1]
)$meta$count
})
## build summary data frame
colnames(basis_occ) <- c("target", "source", "value")
## build links and nodes
links_final <- rbind(links_plot[, 1:3], basis_occ)
links_final <- links_final[links_final$value != 0, ]
nodes_final <- rbind(
nodes_plot,
data.frame(
name = unique(basis_occ$target),
group = "d"
)
)
nodes_final <- nodes_final[nodes_final$name %in% c(links_final$source, links_final$target), ]
links_final$IDsource <- match(links_final$source, nodes_final$name) - 1
links_final$IDtarget <- match(links_final$target, nodes_final$name) - 1
## make sankey
my_color <- 'd3.scaleOrdinal() .range(["#FDE725FF","#1F9E89FF","#440154FF", "#6b0311"])'
SN <- sankeyNetwork(
Links = links_final, Nodes = nodes_final,
Source = "IDsource", Target = "IDtarget",
Value = "value", NodeID = "name",
colourScale = my_color, NodeGroup = "group",
sinksRight = FALSE, nodeWidth = 40, fontSize = 13, nodePadding = 5, margin = 0,
width = 1100, height = 600
)
networkD3::saveNetwork(network = SN, file = "SankeyFinal.html", selfcontained = TRUE)
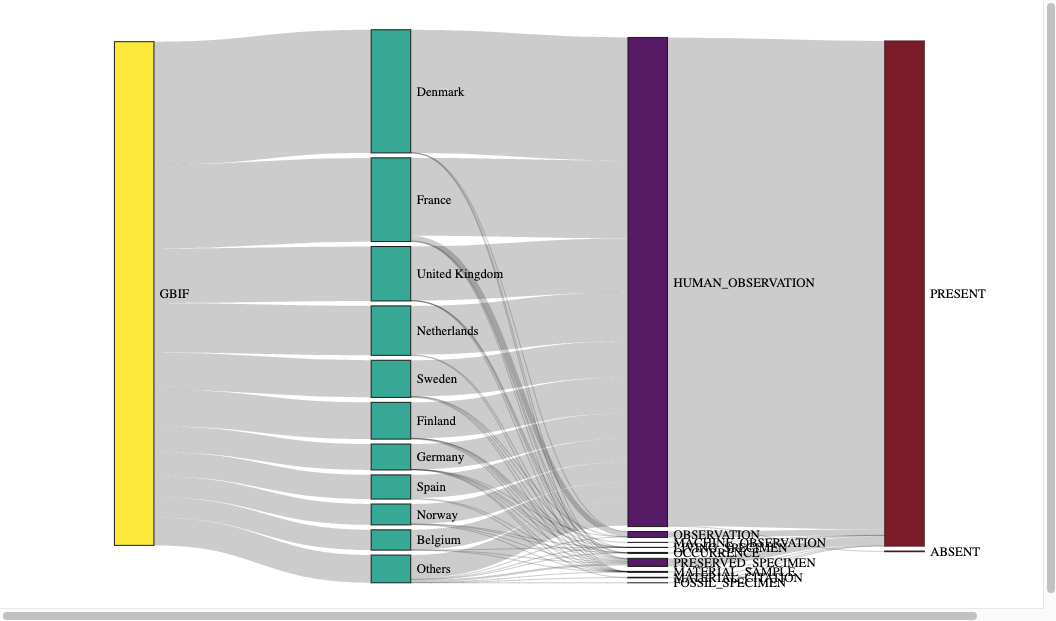
Note for Firefox users: Sankey diagrams are contained in a viewbox which scales poorly on Firefox. You can open this webpage in a different browser to avoid this issue. Alternatively, I have included a .png of this particular diagram in the code-fold above it.
Data Discovery Beyond Counts
The occ_search(...) function is useful for much more than “just” finding out how many GBIF mediated records fit your requirements.
Let me demonstrate the richness of the output returned by occ_search(...) with a simple example of Lagopus muta focussed on Norway:
occ_meta <- occ_search(taxonKey = sp_key, country = "NO")
Now let’s look at the structure of this object:
str(occ_meta, max.level = 1)
## List of 5
## $ meta :List of 4
## $ hierarchy:List of 1
## $ data : tibble [500 × 112] (S3: tbl_df/tbl/data.frame)
## $ media :List of 500
## $ facets : Named list()
## - attr(*, "class")= chr "gbif"
## - attr(*, "args")=List of 6
## - attr(*, "type")= chr "single"
occ_search(...) returns this information:
meta- this is the metadata which we have already used part ofhierarchy- this is the taxonomic hierarchy for thetaxonKey(s) being querieddata- individual data records for our query (a maximum of 100,000 can be obtained perocc_search(...)query)media- media metadatafacets- results can be faceted into individual lists using thefacetargument
We will look at the downloaded data more explicitly in the next section of this workshop.
Additional Data Discovery Considerations
The Darwin Core is large and there are many ways by which to discover different subsets of data left to explore. I leave it up to you, the reader to do so. A good place to start is the documentation for occ_search(...) or the documentation of the
Darwin Core itself:
rgbif. Next, you will need to learn how to actually obtain that data.
Building a Final Data Discovery Query
To facilitate the rest of this workshop, let’s assume we are interested in all records of Lagopus muta across Norway which have been made by humans and found our study organism to be present between and including 2000 and 2020.
occ_final <- occ_search(
taxonKey = sp_key,
country = "NO",
year = "2000,2020",
facet = c("year"), # facetting by year will break up the occurrence record counts
year.facetLimit = 23, # this must either be the same number as facets needed or larger
basisOfRecord = "HUMAN_OBSERVATION",
occurrenceStatus = "PRESENT"
)
knitr::kable(t(occ_final$facet$year))
| name | 2018 | 2019 | 2020 | 2017 | 2016 | 2015 | 2014 | 2012 | 2013 | 2011 | 2010 | 2009 | 2008 | 2007 | 2005 | 2006 | 2004 | 2001 | 2003 | 2000 | 2002 |
| count | 1156 | 1100 | 1021 | 865 | 842 | 841 | 774 | 757 | 691 | 600 | 503 | 354 | 269 | 148 | 114 | 110 | 102 | 95 | 92 | 89 | 83 |
How many records does this return to us? Let’s see:
occ_count(
taxonKey = sp_key,
country = "NO",
year = "2000,2020", # the comma here is an alternative way of specifying a range
basisOfRecord = "HUMAN_OBSERVATION",
occurrenceStatus = "PRESENT"
)
## [1] 10606
We could have also just summed up the facet counts, but it is good to remember this more direct function exists.
Note that we have to change the way we sum the number of records as data discovery for any argument being matched by multiple characteristics generates an output of type list:
str(occ_final, max.level = 1)
## List of 5
## $ meta :List of 4
## $ hierarchy:List of 1
## $ data : tibble [500 × 117] (S3: tbl_df/tbl/data.frame)
## $ media :List of 500
## $ facets :List of 2
## - attr(*, "class")= chr "gbif"
## - attr(*, "args")=List of 10
## - attr(*, "type")= chr "single"
Session Info
## R version 4.4.0 (2024-04-24 ucrt)
## Platform: x86_64-w64-mingw32/x64
## Running under: Windows 11 x64 (build 22631)
##
## Matrix products: default
##
##
## locale:
## [1] C
##
## time zone: Europe/Oslo
## tzcode source: internal
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] pbapply_1.7-2 leaflet_2.2.2 htmlwidgets_1.6.4 networkD3_0.4
## [5] ggplot2_3.5.1 sf_1.0-17 rnaturalearth_1.0.1 knitr_1.48
## [9] rgbif_3.8.1
##
## loaded via a namespace (and not attached):
## [1] gtable_0.3.6 xfun_0.47 bslib_0.8.0 vctrs_0.6.5
## [5] tools_4.4.0 crosstalk_1.2.1 generics_0.1.3 curl_5.2.2
## [9] parallel_4.4.0 tibble_3.2.1 proxy_0.4-27 fansi_1.0.6
## [13] highr_0.11 pkgconfig_2.0.3 R.oo_1.26.0 KernSmooth_2.23-22
## [17] data.table_1.16.0 lifecycle_1.0.4 R.cache_0.16.0 farver_2.1.2
## [21] compiler_4.4.0 stringr_1.5.1 munsell_0.5.1 terra_1.7-78
## [25] codetools_0.2-20 htmltools_0.5.8.1 class_7.3-22 sass_0.4.9
## [29] yaml_2.3.10 lazyeval_0.2.2 pillar_1.9.0 jquerylib_0.1.4
## [33] whisker_0.4.1 R.utils_2.12.3 classInt_0.4-10 cachem_1.1.0
## [37] wk_0.9.4 styler_1.10.3 tidyselect_1.2.1 digest_0.6.37
## [41] stringi_1.8.4 dplyr_1.1.4 purrr_1.0.2 bookdown_0.40
## [45] labeling_0.4.3 fastmap_1.2.0 grid_4.4.0 colorspace_2.1-1
## [49] cli_3.6.3 magrittr_2.0.3 triebeard_0.4.1 crul_1.5.0
## [53] utf8_1.2.4 e1071_1.7-16 withr_3.0.2 scales_1.3.0
## [57] oai_0.4.0 rmarkdown_2.28 httr_1.4.7 igraph_2.1.1
## [61] blogdown_1.19 R.methodsS3_1.8.2 evaluate_0.24.0 s2_1.1.7
## [65] urltools_1.7.3 rlang_1.1.4 Rcpp_1.0.13 httpcode_0.3.0
## [69] glue_1.7.0 DBI_1.2.3 xml2_1.3.6 jsonlite_1.8.8
## [73] R6_2.5.1 plyr_1.8.9 units_0.8-5